Quick Navigation:
Where is the dataset hosted?
How do I apply for access to the ADSP dataset?
What data are available and what’s the expected timeline for future releases?
Is there a cost associated with downloading data?
Can I run programs on S3 without downloading the data?
What are the limitations in using the data?
What are the limitations in exchanging data and information from ADSP with other investigators?
What type of phenotype data do you collect and where can I find additional phenotypes?
Can I request a biospecimen for a sequenced sample I am interested in running additional assays on?
How do I acknowledge the ADSP in my publications or presentations?
How do I contact NIAGADS?
Where is the dataset hosted?
How do I apply for access to the ADSP dataset?
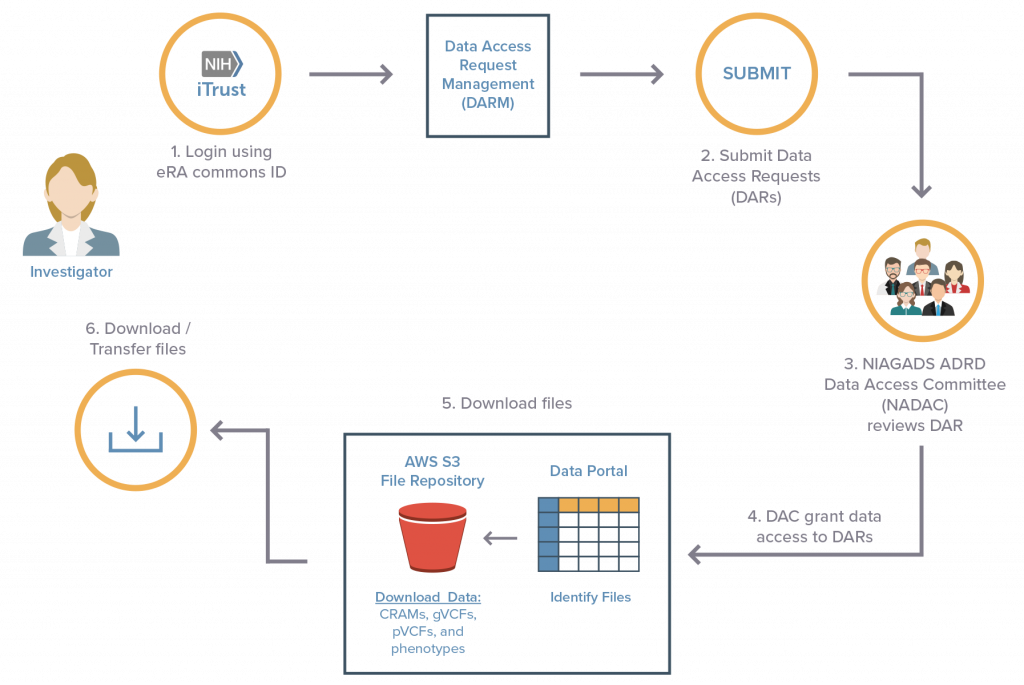
Apply for data access
- The ADSP sequencing data is hosted by NIAGADS DSS.
- Go to the NIAGADS Application Instructions page for application requirements.
- Go to the PI User Guide to find instructions on how to use the application system and data portal
Additional Instruction
- How to use the Data Portal: Portal User Guide
- How to use the Data Access Request Management (DARM) system for data requests: DARM PI User Guide
- Files <5Gb can be downloaded directly through the web portal.
- Files >5Gb must be downloaded directly from Amazon.
- How to set up an Amazon account and download the data: Amazon Instructions
What data are available and what’s the expected timeline for future releases?
Is there a cost associated with downloading data?
NIAGADS incurs the cost for investigators to download most of the ADSP data, including joint genotype-called project level VCFs, phenotypes, and associated meta-data.
CRAMs, gVCFs, and SV VCFs can be downloaded using the Amazon Requester Pays option, which means that the requesting institution will incur the cost of downloading the data.
Options for AWS download using Requestor Pays option:
- AWS resource- You would not be charged if you download within the same region as our S3 bucket, US-East (N. Virginia), to another US-East (N. Virginia)
- Local download- an affordable transfer option is an Amazon Snowball. DSS would send the data to your S3 bucket, then you can create an AWS Snowball export. The device costs $250 to transfer 80TB of data (plus additional fees.
Can I run programs on S3 without downloading the data?
Certain tools can make use of S3 URLs to read data without having to download the file. Our CRAM files can be read by S3-aware alignment reader such as samtools. This allows users to download either a portion or all data from a file without having to save the entire file to a local drive. Although file access may be slower, there is cost savings.
What are the limitations in using the data?
You must have an approved Data Access Request through NIAGADS DSS. The data may only be used in accordance with your approved research use statement and local IRB approval.
What are the limitations in exchanging data and information from ADSP with other investigators?
Internal Investigators
Investigators can share data with other investigators within their institution if (1) all research being conducted is in accordance with the submitting investigators’ approved research use and (2) all investigators accessing the data have read and signed the University of Pennsylvania Data Transfer Agreement. The primary investigator will be responsible for the conduct of anyone accessing the data under their approved DAR.
External Investigators
Investigators can share data with collaborators outside of the PI’s institution. The collaborators must submit parallel project requests with (1) the same project title and (2) the same Research Use Statement and Cloud Use Statement, if applicable, that references the collaboration (for smaller collaborations, the name and institution of the collaborating PI(s) or for larger efforts, the consortium name). Note: for-profit and non-profit institutions may not be able to share all data across sites as some ADSP data cannot be used by for-profit institutions.
What type of phenotype data do you collect and where can I find additional phenotypes?
ADSP Minimal Dataset
Accompanying the sequencing data are a basic set of phenotypes collected from each of the submitting cohorts and harmonized into the ADSP format. There are currently 4 different data dictionaries for the samples included in the ADSP:
Phenotype Harmonization Consortium
Funded in mid-2021, the consortium will harmonize the following endophenotype domains: Cognition, Fluid Biomarkers (CSF), Amyloid PET, Structural MRI (T1), Diffusion MRI, Vascular Risk Factors, and Autopsy (neuropathology). The group plans to conduct yearly releases with the first release occurring in fall of 2022. The exact timeline is contingent on the availability of raw data and the data being of sufficient quality to allow for harmonization.
The first release included a subset of harmonized, ADSP-sequenced data from the Adult Changes in Thought (ACT) Study, Alzheimer’s Disease Neuroimaging Initiative (ADNI), the National Alzheimer’s Coordinating Center (NACC), National Institute on Aging Late Onset of Alzheimer’s Disease (NIA-LOAD), the Religious Orders Study, Memory and Aging Project at Rush, and Minority Aging Research Study (ROS/MAP – Rush/MARS), and the Memory and Aging Project at the Knight ADRC (Knight ADRC). ReadMe files, data dictionaries, and harmonized data files are available across the following domains for joint genomic analysis:
- Cognitive Harmonization (ACT; ADNI; NACC; ROS, MAP – Rush, MARS)
- Fluid Biomarker Harmonization (ADNI; NACC; Knight ADRC)
- Neuropathology Harmonization (NACC; NIA-LOAD, ROS, MAP – Rush, MARS)
NACC
Additional phenotypes for the participants recruited from the Alzheimer’s Disease Research Centers (ADRCs) sequenced by ADSP can be requested directly from the National Alzheimer’s Coordinating Center (NACC).
ADNI
Additional phenotypes for the participants recruited by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) can be requested directly from LONI.
Individual cohorts
Contact the cohort PIs for more information about the phenotypes collected by their cohorts.
RUSH
Additional phenotypes for the participants recruited from the ROS, MAP, or MARs studies can be requested directly from the Rush Alzheimer’s Disease Center (RADC).
Can I request a biospecimen for a sequenced sample I am interested in running additional assays on?
Yes, if the sample is stored at the National Centralized Repository for Alzheimer’s Disease and Related Dementia’s (NCRAD), you can find details and apply for access by visiting this page of the NCRAD website.
How do I acknowledge the ADSP in my publications or presentations?
How do I contact NIAGADS?
For any questions related to the ADSP dataset or how to access it, contact NIAGADS at niagads@pennmedicine.upenn.edu, and they can forward any questions to the appropriate parties.